Oliver Weichenrieder
Structural Biology of Selfish RNA
Max Planck Institute for Biology Tübingen
Adjunct faculty in: IMPRS

Vita
- PhD at the European Molecular Biology Laboratory, Grenoble (1995-1999)
- Postdoctoral Fellow at the Netherlands Cancer Institute, Amsterdam (2000-2004)
- NWO Primary Investigator at the Netherlands Cancer Institute, Amsterdam (2004-2006)
- Departmental Group Leader at the MPI for Developmental Biology (2006-2018)
- Research Group Leader and Head of Genome Center Facility (since 2018)
Research Interest
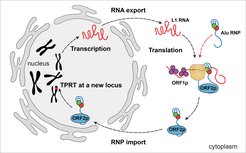
‘Selfish’ RNA likely is at the origin of all life on earth and it persists today in the form of retrotransposons and RNA-based viruses. We study human LINE-1 and Alu RNAs and how these ‘molecular parasites’ copy their sequences into genomic DNA. We are interested in the molecular details that govern this integration process and combine mechanistic analyses based on molecular structures with cell-based retrotransposition assays.
Non-LTR retrotransposons belong to the simplest forms of selfish RNA. These RNAs have no viral relatives and integrate into the genomes of their hosts by target-primed reverse transcription, which occurs directly on genomic DNA and is fundamentally distinct from the cytoplasmic processes mediated by the long terminal repeats of LTR retrotransposons and retroviruses.
Over time, LINE-1 and Alu RNAs have retro-copied themselves into more than 1.5 million genomic locations with consequences for human evolution and disease that are just beginning to unravel. LINE-1 RNA encodes two proteins (L1ORF1p and L1ORF2p) that are essential for its propagation, whereas Alu RNA has evolved as a ‘parasite’ of LINE-1 RNA, which ‘hijacks’ the L1ORF2p reverse transcriptase for its own propagation.
Our current projects therefore aim to shed light on
- the structure and function of the L1ORF1 protein in LINE-1 retrotransposition
- the mechanism of target-primed reverse transcription by L1ORF2p
- the structural features of Alu RNA required to exploit the LINE-1 retrotransposition machinery
In the long run we aim at a more general picture of the ‘molecular ecology’ of selfish RNAs and of their molecular interactions with the respective hosts, as these continue to drive evolutionary innovation in many areas of biology.
Available PhD projects
- Currently not recruiting PhD students
Selected Reading
- Ahl, V., Keller, H., Schmidt, S. and Weichenrieder, O. (2015) Retrotransposition and crystal structure of an Alu RNP in the ribosome-stalling conformation. Molecular Cell, 60, 715-727.
- Khazina, E. and Weichenrieder, O. (2018) Human LINE-1 retrotransposition requires a metastable coiled coil and a positively charged N-terminus in L1ORF1p. eLife, 7, e34960.
- Repanas, K., Zingler, N., Layer, L.E., Schumann, G.G., Perrakis, A. and Weichenrieder, O. (2007) Determinants for DNA target structure selectivity of the human LINE-1 retrotransposon endonuclease. Nucleic Acids Res, 35, 4914-4926.

