Andrei Lupas
Protein Evolution
Max Planck Institute for Biology Tübingen
Faculty in: TIPP, IMPRS
Vita

- PhD in Molecular Biology at Princeton University,1985-91
- Postdoctoral training at the Gene Center of the University of Munich and at the MPI for Biochemistry, Martinsried,1993-97
- Senior Computational Biologist and Assistant Director of Bioinformatics at SmithKline Beecham Pharmaceuticals,1997-2001
- Director at the MPI since 2001
Research Interest
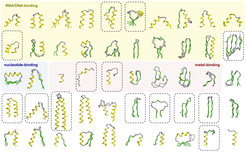
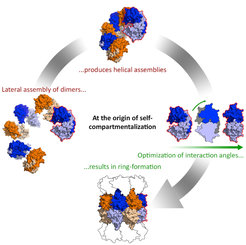
Proteins are essential components of all living cells. It is thus hardly surprising that precursors of most proteins observed today existed at the time of the last common ancestor of all life. How did they evolve? Randomly synthesized polypeptide chains form folded structures in less than one in a billion cases, so it seems impossible that proteins arose by chance. We have proposed that folded proteins evolved by fusion and accretion from an ancestral set of peptides active as cofactors of RNA-dependent replication and catalysis. Using bioinformatics, we have reconstructed this "vocabulary" of ancient peptides and are now exploring experimentally the processes by which it could have led to the emergence of folded proteins, using methods in biochemistry and structural biology (crystallography, NMR, cryo-EM).
We also study how changes in protein structure create new functionality, both by attempting to functionalize newly created proteins in vitro and by exploring structure-function relationships in natural proteins. In the latter, our particular focus is on signal transduction across membranes and cellular mechanisms for protein quality control. An essential aspect of our work is the development of new bioinformatic tools, which we deploy in our MPI Bioinformatics Toolkit.
Available PhD projects
- Currently not recruiting PhD students
Selected Reading
- Zimmermann L, Stephens A, Nam SZ, Rau D, Kübler J, Lozajic M, Gabler F, Söding J, Lupas AN, Alva V. (2018) A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J Mol Biol. 430:2237-43.
- Afanasieva E, Chaudhuri I, Martin J, Hertle E, Ursinus A, Alva V, Hartmann MD, Lupas AN. (2020) Structural diversity of oligomeric β-propellers with different numbers of identical blades. Elife 8:e49853.
- Fuchs AC, Ammelburg M, Martin J, Schmitz RA, Hartmann MD, Lupas AN. (2021) Archaeal Connectase is a specific and efficient protein ligase related to proteasome β subunits. PNAS 118(11).


