Marja Timmermans
miRNAs as Mobile Signals in Development
University of Tübingen
Faculty in: IMPRS

Vita
- PhD studies at Rutgers University, 1990-1996
- Postdoctoral training at Yale University, 1996-1998
- Assistant, Associate, and Full Professor at Cold Spring Harbor Laboratory, 1998-2015
- Alexander von Humboldt Professor at the University of Tübingen since 2015
Research Interest
The focus of our research is on pattern formation in development. We investigate how new tissues and organs arise at plant stem cell niches. We combine molecular genetics with single cell genomic approaches, quantitative time-lapse imaging, and theoretical modeling to discern the spatiotemporal patterns of gene regulation driving cell fate decisions. Please see here for more detail.
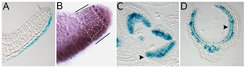
Figure 1. Gradients of mobile small RNAs have morphogen-like patterning activities. Mobility of miRNAs from their site of biogenesis in the bottom epidermis (A) yields a miRNA gradient (B) that through a threshold-based read-out establishes an on-off pattern of target gene expression (C, D).
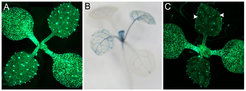
Figure 2. Synthetic system to study miRNA mobility. In seedlings expressing fluorescent reporters (A), artificial miRNAs targeting these reporters are expressed in a regulated often tissue-specific manner (B) to arrive at the parameters and mechanisms of miRNA mobility (C)
Available PhD Projects
- Currently not recruiting PhD students
Selected Reading
- Scacchi et al. (2024) A diffusible small-RNA-based Turing system dynamically coordinates organ polarity. Nat Plants 10, 412–422.
- Burian A., et al. (2022) Specification of leaf dorsiventrality via a prepatterned binary readout of a uniform auxin input. Nat Plants 8, 269-280.
- Denyer, T., et al. (2019) Spatiotemporal developmental trajectories in the Arabidopsis root revealed using high-throughput single cell RNA sequencing. Dev Cell 48, 840-852.
- Skopelitis, D., et al. (2018) Gating of miRNA movement at defined cell-cell interfaces governs their impact as positional signals. Nat Commun. 9, 3107.
- Skopelitis, D., et al. (2017) Boundary formation through a direct threshold-based readout of mobile small RNA gradients. Dev Cell 43, 265-273.


