Gal Ofir
Plant Immunogenomics
Max Planck Institute for Biology Tübingen
Adjunct faculty in: IMPRS

Vita
- PhD, Weizmann Institute of Science, Israel (2015-2021)
- EMBO & HFSP Postdoctoral Research Fellow, Department of Molecular Biology, Max Planck Institute for Biology Tübingen, Germany (2022-2024)
- Project Leader, Department of Molecular Biology, Max Planck Institute for Biology Tübingen, Germany (2025-present)
Research Interest
The evolution of immunity and acute stress-tolerance is unique in its rapid pace and innovative nature. As organisms co-evolve with pathogens and parasites, both sides constantly have to change and adapt to outsmart the other. This leads to fantastic innovation of molecular immune systems across the tree of life. At the same time, the race between parasites and hosts is as ancient as life itself; It was only recently appreciated that many of the immune mechanisms we observe in animals and plants today have their roots in our original common ancestors – prokaryotic cells that were defending themselves against viruses since the dawn of cellular life on Earth.
In our group, we aim to learn from discoveries in immunity and rapid evolution across the tree of life to decipher the complex ways by which plants adapt to pathogen and environmental stresses. Our research focus on both extreme ends of evolutionary dynamics to discover new genes involved in adaptation and immunity:

1. Diversification - rapidly changing selection pressure shape rapidly evolving genomic loci. These genomic regions are characterized by novel genes, rampant gene duplication and translocation, and hotspots of mobile genetic elements' activity. This makes them the hardest regions to study in complex plant genomes. Recent technological developments finally allow us to clearly see and compare these islands of diversity in large collection of plants from the same species. We are utilizing these new resources to discover new gene families that exhibit high diversity and rapid evolution to identify new candidate genes that could be involved in immunity and adaptation to stress.
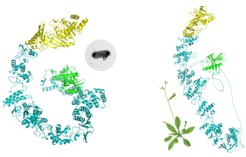
2. Conservation - remarkable conservation of immune modules can be observed across immune systems of animals, bacteria and plants. Immunity shares a common language of protein domains, a language we are finally getting to know. We use this conservation and reuse of immune proteins domains to identify plant homologues of bacterial and animal immune systems, using the vast knowledge of immunity in other organism to better understand plant immunity.
Available PhD Projects
- Currently not recruiting PhD students
Selected Reading
- Gal Ofir, Ehud Herbst, Maya Baroz, Daniel Cohen, Adi Millman, Shany Doron, Nitzan Tal, Daniel B. A. Malheiro, Sergey Malitsky, Gil Amitai, Rotem Sorek. Antiviral activity of bacterial TIR domains via signaling molecules. Nature 600, 116-120 (2021)
- Jean Cury, Matthieu Haudiquet, Veronica Hernandez Trejo, Ernest Mordret, Anael Hanouna, Maxime Rotival, Florian Tesson, Delphine Bonhomme, Gal Ofir, Lluis Quintana-Murci, Philippe Benaroch, Enzo Z Poirier, Aude Bernheim. Conservation of antiviral systems across domains of life reveals novel immune mechanisms in humans. Cell Host & Microbe 32, 1594-1607 (2024)
- Adi Millman, Sarah Melamed, Azita Leavitt, Shany Doron, Aude Bernheim, Jens Hör, Jeremy Garb, Nathalie Bechon, Alexander Brandis, Anna Lopatina, Gal Ofir, Dina Hochhauser, Avigail Stokar-Avihail, Nitzan Tal, Saar Sharir, Maya Voichek, Zohar Erez, Jose Lorenzo M Ferrer, Daniel Dar, Assaf Kacen, Gil Amitai, Rotem Sorek. An expanded arsenal of immune systems that protect bacteria from phages. Cell Host Microbe 30, 1556-1569 (2022)
- Shany Doron, Sarah Melamed, Gal Ofir, Azita Leavitt, Anna Lopatina, Mai Keren, Gil Amitai, Rotem Sorek. Systematic discovery of new anti-phage defense systems in the microbial pan-genome. Science 359, 1008 / eaar4120 (2018)
- Gal Ofir, Sarah Melamed, Hila Sberro, Zohar Mukamel, Shahar Silverman, Gilad Yaacov, Shany Doron, Rotem Sorek. DISARM is a widespread bacterial defence system with broad anti-phage activities. Nature Microbiology 3, 90–98 (2018)


